Deep and Lightweight Neural Network for Histopathological Image Classification
Shin Kim and Kyoungro Yoon*
Konkuk University, Seoul, Republic of Korea
E-mail: yoonk@konkuk.ac.kr
*Corresponding Author
Received 04 April 2021; Accepted 24 February 2022; Publication 07 July 2022
Abstract
Breast cancer is a fatal disease affecting women, and early detection and proper treatment are crucial. Classifying medical images correctly is the first and most important step in the cancer diagnosis stage. Deep learning-based classification methods in various domains demonstrate advances in accuracy.
However, as deep learning improves, the layers of neural networks get deeper, raising challenges, such as overfitting and gradient vanishing. For instance, a medical image is simpler than an ordinary one, making it vulnerable to overfitting issues.
We present breast histopathological classification methods with two deep neural networks, Xception and LightXception with aid of voting schemes over split images. Most deep neural networks classify thousands classes of images, but the breast histopathological image classes are far fewer than those of other image classification tasks. Because the BreakHis dataset is relatively simpler than typical image datasets, such as ImageNet, applying the conventional highly deep neural networks may suffer from the aforementioned overfitting or gradient vanishing problems. Additionally, highly deep neural networks require more resources, leading to high computational costs. Consequently, we propose a new network; LightXception by cutting off layers at the bottom of the Xception network and reducing the number of channels of convolution filters. LightXception has only about 35% of parameters compared to those of the original Xception network with minimal expense on performance. Based on images with 100X magnification factor, the performance comparisons for Xception vs. LightXception are 97.42% vs. 97.31% on classification accuracy, 97.42% vs. 97.42% on recall, and 99.26% vs. 98.67% of precision.
Keywords: Breast cancer classification, image classification, BreakHis dataset, lightweight network, medical image.
1 Introduction
Breast cancer is a common malignancy in women. [1] reports a 5-year related survival of breast cancer of 90% and death rate of 20%. Early discovery of cancer is significant to treating the cancer in proper time and preventing deaths.
Therefore, it is important to classify medical images correctly. In the past few years, machine learning has been adapted and advanced in various domain, such as medical image classification, health monitoring, and more. Before deep learning, studies in medical image classification used traditional feature extractors, but currently, most of the researches utilize deep learning-based classifiers for its superior performance.
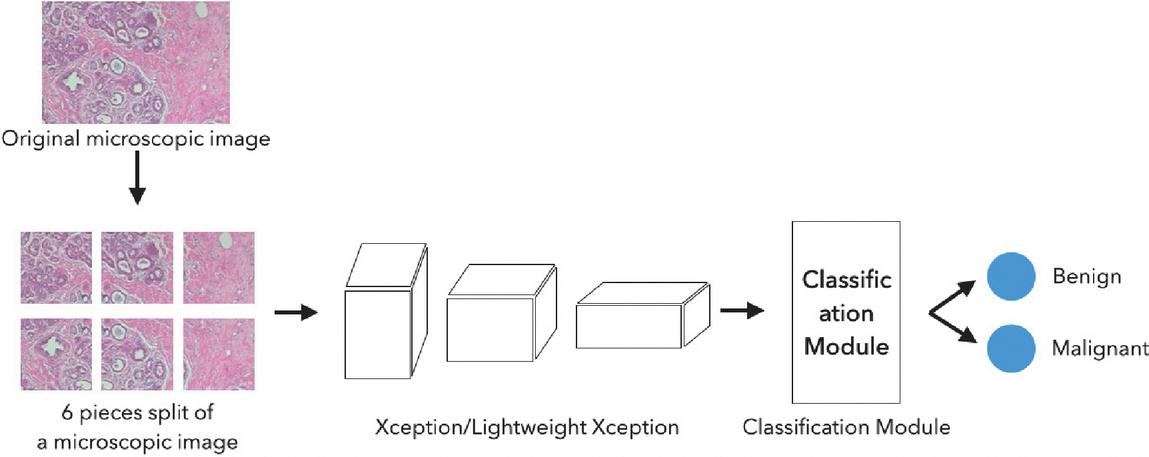
We propose deep neural networks for breast cancer classification using BreakHis dataset. We used Xception network as the base network to identify the type of tumor. Most deep neural networks categorize numerous images into thousands of classes. However, because medical image datasets have a smaller number of images with fewer classes compared with other image data sets, we may not need the complex network, such as the original Xception network, for medical image classifications. Therefore, we lightened the Xception network, named LightXception network, by removing layers at the bottom of the neural network and reducing the number of channels of the convolution filters. LightXception has about 7.9 million parameters, while Xception has about 22.1 million parameters, which is only about 35% of the size of Xception. However, the classification accuracy of breast cancer images is not significantly degraded. We also propose a voting scheme based on split image classification, in which a cancer image is divided into six split images and the final class decision is made based on the results of six split images.
The remainder of this paper is organized as follows. In Section 2, related work of our research is described. Section 3 states our proposed network for breast classification. Then the evaluation method and results are shown in Section 4 and the paper is concluded in Section 5.
2 Related Work
2.1 Medical Image Classification
Image classification algorithms have advanced greatly in the last decade. AlexNet [2] is the first image classification algorithm to use GPU to achieve faster parameter calculation compared with previous image classification algorithms, and triggered the development of networks with lengthy hidden layers, such as [3, 4, 5, 6, 7]. In [3], C. Szegedy et al. present an inception v3 module that builds the neural network deeper, using 1x1 convolution to prevent overfitting and vanishing gradient problems. Xception network [4] is a network based on an inception module with a deeper network and increased parameters. For the Xception network, the inception module was replaced with a separate depthwise convolution comprising a depth-wise and a pointwise convolution. A depthwise convolution is an independent spatial convolution for each channel, and pointwise convolution is a 1x1 convolution that projects into a new channel space. Table 1 illustrates the comparison of classification performances and Table 2 shows the comparison of the number of parameters in Inception V3 and Xception networks. These comparisons demonstrate Xception outperforms Inception V3 with fewer parameters.
Table 1 Classification performance comparison [4]
| Top-1 Accuracy | Top-5 Accuracy | |
| VGG-16 | 0.715 | 0.901 |
| Inception V3 | 0.782 | 0.941 |
| Xception | 0.790 | 0.945 |
Table 2 Size comparison [4]
| Parameters Counts | |
| Inception V3 | 23,626,728 |
| Xception | 22,855,952 |
Based on image classification, medical image classification has been investigated to improve classification performance and to provide medical services remotely. [11] surveys deep learning-based methods for lung nodule classification and reports that [12] and [13] show the best performance on lung nodule classification with [12] achieving 88.96% and [13] 89.99% accuracy.
Most deep neural networks are developed in an environment where GPU resource is unlimited. Since the actual devices using neural networks have more restricted resources compared with the development environment, neural network applicable in restricted environments, such as mobile phones have been investigated.
In [8], J. Shihadeh et al. propose a skin cancer image classification based on AlexNet [2] and GoogLeNet [19] for remote medical diagnosis. The application developed on a light computer node, Nvidia Jetson TX2, achieved 74.57% accuracy. In [9], H. W. Huang et al. present a lightweight skin cancer classification network for cloud application and remote medical services based on EfficientNet [10], achieving 72.1% accuracy.
2.2 Researches Related to BreakHis Dataset
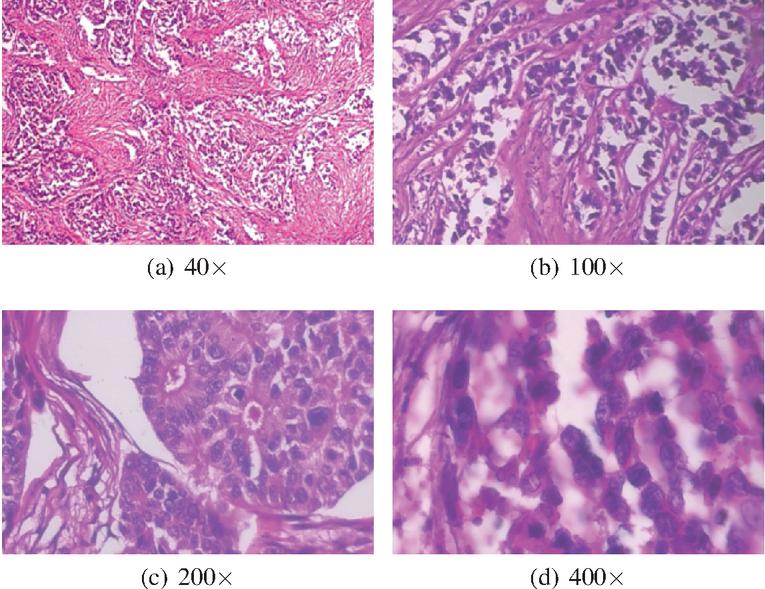
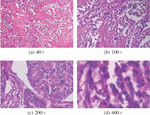
In [14], F. A. Spanhol et al. publish the BreakHis (Breast Cancer Histopathological Image Classification) dataset, composed of 9,109 microscopic images of breast tumor tissue collected from 82 patients using varying magnifying factors of 40, 100, 200 and 400. The dataset includes 2,480 benign and 5,429 malignant samples and its image resolution is 700460 pixels. The dataset comprises two main groups; benign and malignant tumors. The BreakHis data is structured as illustrated in Table 3 and Figure 1 illustrates samples of BreakHis dataset with different magnification factors.
Table 3 BreakHis dataset composition
| Magnification | Benign | Malignant | Total |
| 40 | 652 | 1,370 | 1,995 |
| 100 | 644 | 1,437 | 2,081 |
| 200 | 623 | 1,390 | 2,013 |
| 400 | 588 | 1,232 | 1,820 |
| Total of Images | 2,480 | 5,429 | 7,909 |
Figure 1 Samples of BreakHis dataset [14].
In [14], F. S. Spanhol et al. research breast histopathological image classification for the first time, based on BreakHis dataset. They study breast cancer image classification by utilizing traditional descriptors and classifiers. Further, [15, 16], F. S. Spanhol et al. replace traditional descriptors and classifiers with a deep feature extractor and deep neural network, DeCAF feature and AlexNet, for image classification. Moreover, several institutes conducted studies [18, 20, 21, 24] on breast cancer image classification based on the BreakHis dataset. In [18], B. Wei et al. present a BiCNN network based on GoogLeNet [19], acquiring classification accuracy of 97.89% for 40, 97.64% for 100, 97.56% for 200 and 97.97% for 400. In [20] A. A. Nahid et al. propose classification models based on CNN, which have the capability of extracting features and LSTM, which can take advantage of long-term dependencies of the data sequences. The model combining CNN and LSTM shows the classification accuracies of 84.33% for 40, 86% for 100, 85% for 200 and 85.71% for 400. In [21] X. Li et al. suggested breast cancer image classification using interleaved DenseNet [22] with SENet [23] achieving accuracy performance of 87.1% for 40, 81.9% for 100, 84.4% for 200 and 84% for 400. Kassani S. H. et al. present an ensemble model of multiple CNNs, VGG-16 [25], MobileNet [26] and DenseNet [22] for breast cancer image classification in [24] achieving classification performance of 98.13% accuracy, 98.75% precision, 98.54% recall and 98.64% F1 score.
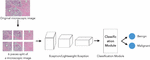
Figure 2 Overall breast cancer classification process with proposed neural networks.
3 Methodology
3.1 LightXception – Lightening the Network
[26, 27] are developed for mobile architecture by making the architecture thinner, extremely reducing the number of parameters of neural networks. Accuracy decrease due to extremely reduced parameters is inevitable, but this decease in accuracy has been minimized by introducing up-to-date concepts, such as depth-wise separable convolution and inverted residuals. This is the main reason we utilize Xception—a neural network for breast cancer classification—rather than a mobile neural network, which is more focused on speed and efficiency over accuracy. Misclassification could be deadly for patients.
Therefore, we propose a LightXception network based on the Xception network for breast cancer histopathological image classification. The LightXception network is defined by removing layers at the bottom of the network and reducing the number of channels of convolution filters. Xception network was developed for image classification, targeting the ImageNet dataset, which comprises thousands of classes of images. To categorize these classes of images, an elaborate network is necessary. However, the BreakHis dataset [14] comprises relatively lesser number of microscopic images, far fewer than the general image dataset. The network is lightened to prevent overfitting and gradient vanishing challenges while saving computing resources.
3.2 Fine-tuning the Networks
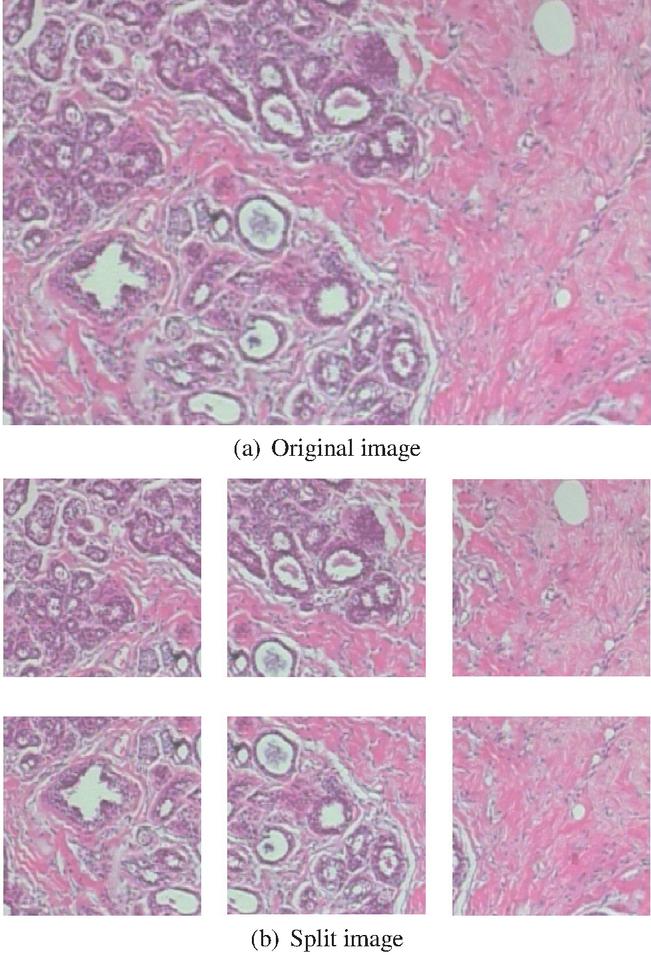
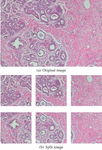
Both the original Xception and LightXception are fine tuned to achieve higher classification performance using Keras Framework [28]. The BreakHis dataset was divided into 5 groups; 4 training groups and a group for evaluation. The neural network was fine-tuned 5 times, that is, a 5-fold validation. Since the microscopic image size of the BreakHis dataset is 700460 pixels, it is essential to resize the image to match the network’s required input image size. However, downscaling the image may cause information loss. Therefore, the image is split into 6 pieces with the resolution of 299299 pixels to match the input size of the network. The split images are overlapped with each other due to the image resolution. Figure 3 is an illustration of an original image and consequent split images.
Figure 3 Samples of original image and split images.
Table 4 Parameters for fine-tuning the networks
| Batch Size | 32 |
| Optimizer | AdaDelta |
| Learning Rate | 0.9 |
| Class Weight | {3.0, 1.0} |
| Loss Function | Huber loss |
| Early Stopping | 250 |
Table 4 illustrates the parameters for fine-tuning the networks. The existing ImageNet weights are used as it is difficult to find the optimal weight of the neural network from all zero weights. ImageNet weights are used for both of the original Xception and LightXception by reshaping the ImageNet weights in line with the LightXception network architecture. Additionally, the class weight is set to 3.0 for benign tumor and 1.0 for malignant tumor as the number of benign tumor images is fewer than malignant tumor images, as illustrated in Table 1. Early stopping is applied with an extra 250 epochs to prevent overfitting challenges. Also, data was augmented using parameters illustrated in Table 5 with the data augmentation generator provided by the Keras Framework.
Table 5 Data augmentation parameters
| Horizontal Flip | True |
| Vertical Flip | True |
| Fill Mode | “nearest” |
| Width Shift Range | 0.2 |
| Height Shift Range | 0.2 |
| Zoom Range | 0.2 |
| Rotation Range | 180 |
3.3 Voting Scheme with Split Images
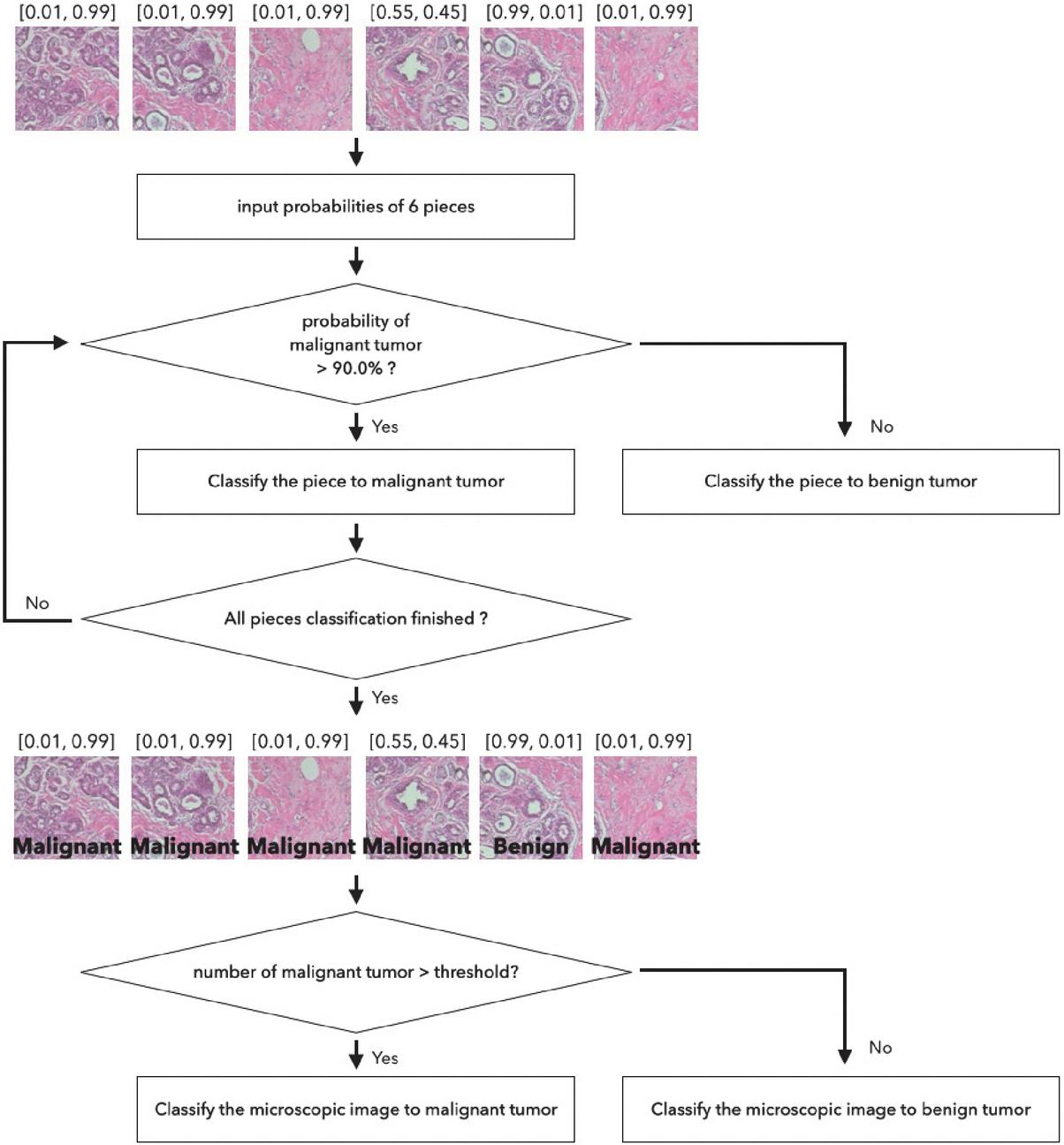
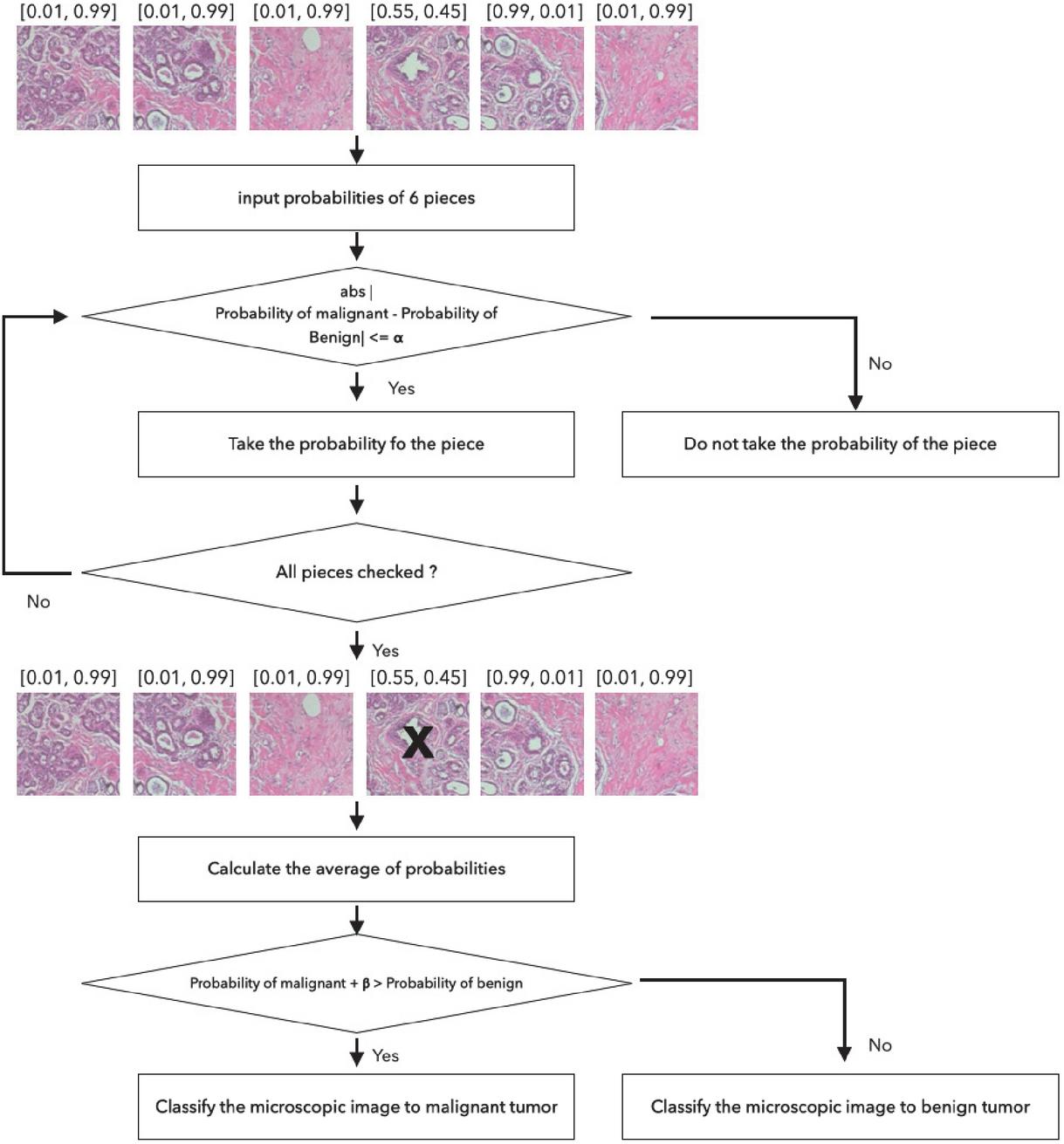
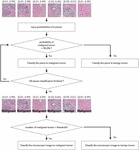
The final decision on the tumor type is made based on the six network outputs for each split image. Each microscopic image is split into six small images, with a resolution of 299299 pixels, with an overlap allowance as aforementioned. Then each split image is fed into the classification network and the probability of breast tumor classes, which are for benign and malignant tumor, acquired in the format of [benign probability, malignant probability]. Figures 4 and 5 illustrate the two voting schemes we are proposing. Figure 4 shows a classification method that considers a threshold value. The probability of breast tumor type for each piece of image is obtained from the networks. If the probability of malignant is higher than 90%, then the tumor is classified as malignant tumor class. If the count of pieces classified as malignant is larger than the threshold value, then the image is classified as malignant tumor image. In this experiment, the best performance is acquired for the threshold value of 3.
Figure 4 Classification module pipeline by using threshold value.
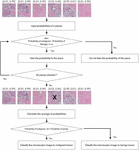
Figure 5 demonstrates a classification method of averaging the probabilities. Similar to the threshold method, a microscopic image is split into 6 pieces and each piece is put into the classification network and probabilities of breast cancer types are acquired. If absolute difference between the probabilities of benign and malignant tumor type is less than for a certain piece of image, the piece is obscure and is not used for classification. The probabilities for malignant and benign types are averaged separately over the remaining pieces. If the averaged probability for malignant class plus , which is a control weight introduced because of the imbalanced dataset, is higher than the averaged probability for the benign class, the image is classified as a malignant tumor class. was set to 0.5 and to 0.1 for evaluation.
Figure 5 Classification module pipeline by calculating average.
4 Evaluation
4.1 Whole Image Analysis
The evaluation of the networks was performed on the full image analysis to check the validity of the voting scheme with split images for Xception and LightXception using measures of precision, recall, accuracy and F1 score, which are calculated with TP (True positive), TN (True negative), FP (False positive), and FN (False negative). TP represents a classified positive when it is actually positive and TN a classified negative when actually it is negative. FP represents that is classified positive, but the answer is negative and FN indicates that is classified negative but is positive. In other words FP and FN are errors that are mis-classified.
| (1) | |
| (2) | |
| (3) | |
| (4) |
In case of full images, an image is classified as a malignant tumor if the predicted probability of the malignant type is higher than 90%. Tables 6 and 7 illustrate classification performance results for full histopathological images.
Table 6 Evaluation Results on original Xception for whole image
| Magnification | Precision | Recall | Accuracy | F1 Score |
| 40 | 99.78 | 99.78 | 99.70 | 99.78 |
| 100 | 99.65 | 99.04 | 99.10 | 99.35 |
| 200 | 99.86 | 99.14 | 99.30 | 99.49 |
| 400 | 99.84 | 99.49 | 99.34 | 99.51 |
Table 7 Evaluation Results on LightXception for whole image
| Magnification | Precision | Recall | Accuracy | F1 Score |
| 40 | 97.85 | 97.23 | 96.59 | 97.51 |
| 100 | 93.14 | 90.81 | 88.41 | 91.68 |
| 200 | 97.51 | 94.53 | 94.49 | 95.82 |
| 400 | 96.76 | 95.54 | 95.82 | 96.12 |
4.2 Split Images-based Classification
Tables 8 and 9 illustrate evaluation results based on Xception and LightXception using the averaging voting scheme, respectively. Tables 10 and 11 indicate evaluation results using the threshold voting scheme. The evaluation results demonstrate Xception obtains similar evaluation results regardless of classification module used, but LightXception has higher classification performance when using a classification module.
Table 8 Evaluation Results on Xception for split images with voting scheme (Averaging)
| Magnification | Precision | Recall | Accuracy | F1 Score |
| 40 | 99.86 | 98.69 | 99.00 | 99.26 |
| 100 | 99.26 | 97.42 | 97.42 | 98.32 |
| 200 | 98.99 | 98.92 | 98.56 | 98.95 |
| 400 | 99.36 | 99.11 | 98.96 | 99.23 |
Table 9 Evaluation Results on LightXception for split images with voting scheme (Averaging)
| Magnification | Precision | Recall | Accuracy | F1 Score |
| 40 | 99.10 | 96.35 | 96.89 | 97.70 |
| 100 | 98.67 | 97.42 | 97.31 | 98.04 |
| 200 | 97.87 | 97.91 | 97.07 | 97.88 |
| 400 | 98.59 | 96.10 | 96.43 | 97.33 |
Table 10 Evaluation Results on Xception for split images with voting scheme (Threshold)
| Magnification | Precision | Recall | Accuracy | F1 Score |
| 40 | 99.86 | 98.76 | 99.05 | 99.30 |
| 100 | 99.00 | 97.84 | 97.84 | 98.40 |
| 200 | 98.92 | 98.99 | 98.56 | 98.96 |
| 400 | 99.44 | 99.11 | 99.01 | 99.27 |
Table 11 Evaluation Results on LightXception for split images with voting scheme (Threshold)
| Magnification | Precision | Recall | Accuracy | F1 Score |
| 40 | 98.15 | 98.54 | 97.69 | 98.33 |
| 100 | 97.87 | 98.19 | 97.26 | 98.02 |
| 200 | 97.26 | 98.87 | 96.87 | 97.87 |
| 400 | 97.97 | 97.16 | 96.70 | 97.55 |
Table 12 Performance differences between original Xception and LightXception (Averaging)
| Magnification | Precision | Recall | Accuracy | F1 Score |
| 40 | 0.75 | 2.34 | 2.11 | 1.56 |
| 100 | 0.59 | 0.00 | 0.12 | 0.28 |
| 200 | 1.12 | 1.01 | 1.49 | 1.08 |
| 400 | 0.77 | 3.00 | 2.53 | 1.90 |
Table 13 Performance differences between original Xception and LightXception (Threshold)
| Magnification | Precision | Recall | Accuracy | F1 Score |
| 40 | 1.71 | 0.22 | 1.36 | 0.97 |
| 100 | 1.13 | 0.35 | 0.158 | 0.38 |
| 200 | 1.74 | 0.12 | 1.69 | 1.09 |
| 400 | 1.47 | 1.95 | 2.32 | 1.72 |
These evaluation results demonstrate that a similar Xception network performance is achievable with LightXception, which has only a little bit over 35% of the number of parameters of Xception network.
Table 14 Breast classification performance comparison
| Magnification Factor | ||||
| Method | 40 | 100 | 200 | 400 |
| Spanhol et al. [15] | 89.5 | 85.0 | 84.0 | 80.8 |
| Spanhol et al. [16] | 84.6 | 84.8 | 84.2 | 81.6 |
| Wei, B. et al. [18] | 97.8 | 97.6 | 97.6 | 98.0 |
| Li, X. et al. [21] | 89.1 | 85.0 | 87.0 | 84.5 |
| Kassani, S. H et al. [24] | – | – | 98.8 | – |
| Xception | 99.0 | 97.4 | 98.6 | 99.0 |
| LightXception | 96.9 | 97.3 | 97.1 | 96.4 |
In addition, as illustrated in Table 11, LightXception provides one of the top performances in breast histopathological image classification.
5 Conclusion and Future Work
We propose a breast histopathological image classification neural network based on Xception, one of the best performing classification neural network. We evaluated the classification performance of original Xception and the proposed LightXception, in which we reduced the number of parameters to 35% of the original Xception by reducing the number of layers and convolution channels.
As a trade-off to the reduction of parameters, the accuracy of LightXception is decreased by not greater 1.9% based on the F-1 score. Moreover, the proposed network, when applied with a voting scheme, shows excellent performance compared with previous research results.
However, there are more lightweight neural networks like the MobileNet and ShuffleNet, which are lighter than the proposed neural network. We plan to utilize these mobile networks to further reduce parameters without degradation. We will expand the classification class from binary to several sub-classes as this will be helpful in providing additional information for treatment.
Acknowledgement
This paper was supported by Konkuk University in 2021.
References
[1] https://seer.cancer.gov/statfacts/html/breast.html
[2] A. Krizhevsky, I. Sutskever and G. E. Hinton, G. E. Imagenet classification with deep convolutional neural networks. In neural information processing systems, 25:1097–1105, 2012
[3] C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, J and Z. Wojna. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition 2818-2826, 2016.
[4] F. Chollet. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition, 1251–1258, 2017
[5] K. He, X. Zhang, S. Ren, and Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, 770–778, 2016.
[6] C. Szegedy, S. Ioffe, V. Vanhoucke, and V. Alemi. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, 31(1), 2017
[7] S. Xie, R. Girshick, P. Dollár, Z. Tu, and K. He. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 1492–1500, 2017.
[8] J. Shihadeh, A. Ansari and T. Ozunfunmi. Deep learning based image classification for remote medical diagnosis. In 2018 IEEE Global Humanitarian Technology Conference (GHTC), 1–8, 2018
[9] H. W. Huang, B. W. Y. Hsu, C. H. Lee and V. S. Tseng. Development of a light‐weight deep learning model for cloud applications and remote diagnosis of skin cancers. In The Journal of Dermatology, 48(3):310–316, 2021
[10] M. Tan and Q. Le. Efficientnet: Rethinking model scaling for convolutional neural networks. In International Conference on Machine Learning, 6105–6114, 2019
[11] P. K. Illa, T. S. Kumar and F. S. A. Hussainy. Deep Learning Methods for Lung Cancer Nodule Classification: A Survey. In Journal of Mobile Multimedia, 18(2):421–450, 2021
[12] X. Xu, C. Wang, J. Guo, L. Yang, H. Bai, W. Li, and Z. Yi. DeepLN: a framework for automatic lung nodule detection using multi-resolution CT screening images. In Knowledge-Based Systems, 189:105–128, 2020
[13] M. Tan, F. Wu, B. Yang, J. Ma, D. Kong, Z. Chen and D. Long. Pulmonary nodule detection using hybrid two‐stage 3D CNNs. In Medical physics, 47(8):3376–3388, 2020
[14] F. A. Spanhol, L. S. Oliveira, C. Petitjean and L. Heutte. A dataset for breast cancer histopathological image classification. In IEEE transactions on biomedical engineering, 63(7):1455–1462, 2015.
[15] F. A. Spanhol, L. S. Oliveira, C. Petitjean and L. Heutte. Breast cancer histopathological image classification using convolutional neural networks. In 2016 international joint conference on neural networks (IJCNN), 2560–2567, 2016.
[16] F. A. Spanhol, L. S. Oliveira, P. R. Cavalin, C. Petitjean and L. Heutte. Deep features for breast cancer histopathological image classification. In 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), 1868–1873, 2017.
[17] J. Donahue, Y. Jia, O. Vinyals, J. Hoffman, N. Zhang, E. Tzeng and T. Darrell. Decaf: A deep convolutional activation feature for generic visual recognition. In International conference on machine learning, 647–655, 2014
[18] B. Wei, Z. Han, X. He and Y. Yin. Deep learning model based breast cancer histopathological image classification. In 2017 IEEE 2nd international conference on cloud computing and big data analysis (ICCCBDA), 348–353, 2017.
[19] C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov and A. Rabinovich. Going deeper with convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition, 1–9, 2015.
[20] A. A. Nahid, M. A. Mehrabi and Y. Kong. Histopathological breast cancer image classification by deep neural network techniques guided by local clustering. In BioMed research international, 2018.
[21] X. Li, X. Shen, Y., Zhou, X. Wang, and T. Q. Li. Classification of breast cancer histopathological images using interleaved DenseNet with SENet (IDSNet). In PloS one, 15(5), 2020.
[22] G. Huang, Z. Liu, L. Van Der Maaten and. K. Q. Weinberger. Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 4700–4708, 2017.
[23] J. Hu, L, Shen and G. Sun. Squeeze-and-excitation networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 7132–7141, 2018
[24] S. H. Kassani, P. H. Kassani, M. J. Wesolowski, K. A. Schneider, K. A and R. Deters. Classification of histopathological biopsy images using ensemble of deep learning networks. In arXiv preprint arXiv:1909.11870, 2019.
[25] K. Simonyan and A. Zisserman. Very deep convolutional networks for large-scale image recognition. In arXiv preprint arXiv:1409.1556, 2014.
[26] A. G. Howard, M. Zhu, B. Chen, D. Kalenichenko, W. Wang, T. Weyand and H. Adam. Mobilenets: Efficient convolutional neural networks for mobile vision applications. In arXiv preprint arXiv:1704.04861, 2017.
[27] M. Sandler, A. Howard, M. Zhu, A. Zhmoginov and L. C. Chen, L. C. Mobilenetv2: Inverted residuals and linear bottlenecks. in Proceedings of the IEEE conference on computer vision and pattern recognition, 4510–4520, 2018
[28] https://keras.io
Biographies

Shin Kim received the B.S degree in computer science engineering from Konkuk University, Seoul, Republic of Korea, the Master degree in computer science engineering from Konkuk University, Seoul, Republic of Korea in 2017. She is a Ph. D. student in computer science engineering at Konkuk University. Her research interests include artificial intelligence, deep learning, image processing and standardization.

Kyoungro Yoon received the BS degree in computer and electronic engineering from Yonsei University, Seoul, Republic of Korea in 1987, the MSE degree in electrical engineering/systems from the University of Michigan, Ann Arbor in 1989, and the Ph.D. in computer and information science from Syracuse University in 1999. He was a principal researcher and a group leader in the Mobile Multimedia Research Lab, LG Electronics Institute of Technology from 1999 to 2003. He joined the school of Computer Science and Engineering of Konkuk University, Seoul, Korea in 2003 as an assistant professor and became a full professor in 2012. He has been with the department of Smart ICT Convergence, since 2017. He has also served as a co-chair of the Ad Hoc Group on User Preferences and the chair of the Ad Hoc Group on MPEG Query Format and Ad Hoc Group on MPEG-V of ISO/IEC JTC1 SC29 WG11 (MPEG). He also served as the chair of the Metadata Subgroup and JPSearch Ad Hoc Group of ISO/IEC JTC1 SC29 WG1 (i.e., JPEG). He is an editor of various international standards, such as ISO IS 15938-12, 23005-1, 23005-2, 23005-5, 23005-6, 23093-1, 24800-3, 24800-5, and 24800-6. He currently serves as the chair of IEEE-SA 2888 WG. His main research interests include smart media systems, image processing, multimedia information and metadata processing.
Journal of Mobile Multimedia, Vol. 18_6, 1913–1930.
doi: 10.13052/jmm1550-4646.18619
© 2022 River Publishers