Modified DNA-based Cryptography System in the Cloud: Deep Maxout-based Fined Tuned Key Generation
Garima Verma
School of Computing, DIT University, Dehradun, India
E-mail: garimaverma.research@gmail.com
Received 12 January 2023; Accepted 29 December 2023; Publication 22 February 2024
Abstract
Cloud security is a set of practices and tools created to address both internal and external security threats to businesses. Organizations must have cloud security as they implement their digital transformation schemes and include cloud-based tools and services in their infrastructure. Cryptography is a mechanism for preventing illegal access to data. In this paper, modified DNA-based cryptography (MDNAC), which is defined as data hiding with respect to DNA sequence is used. The steps involved in the proposed MDNAC is: encryption and decryption with optimal key generation. A way of converting plain text into cipher text is known as encryption. Two components make up the encryption process: a key and an encryption algorithm. For the encryption algorithm, we employed a modified DNA algorithm. In the decryption phase, the reverse operation is performed to get the plain text from the cipher text. Moreover, a deep learning model is used for generating the keys; the model used is deep maxout. To ensure the appropriate key generation process, the weights of the deep maxout are optimally tuned by the new feedback assisted Archimedes optimization (FAAO) algorithm. Based on the generated keys, the encryption process takes place. Finally, the performance of MDNAC is evaluated using conventional methods with respect to different measures. Additionally, the MDNAC obtained a correlation value of 0.20297 for the mean case scenario, despite the fact that the corresponding values are 0.02%, 0.17%, 0.2%, 0.7%, 0.12%,0.41%, 0.86%, and 0.46% as compared to the other models such as FAT, AOA, BMO, COOT, BOA, SSO, WOA, and LES respectively.
Keywords: Cloud security, cryptography, encryption, DNA, deep learning.
Nomenclature
| Abbreviation | Description |
| OTP | One-time pad |
| IoT | Internet of Things |
| DNA | Deoxyribonucleic acid |
| CDMB | Central dogma of molecular biology |
| BAMNN | Bidirectional associative memory neural network |
| WOA | Whale optimization algorithm |
| RSA | Rivest Shamir Adleman |
| MDNAC | Modified DNA based cryptography |
| FAAO | Feedback assisted Archimedes optimization algorithm |
| DMN | Deep maxout algorithm |
1 Introduction
Cloud security, often referred to as cloud computing security, is a group of security controls intended to safeguard data, apps, and infrastructure that are stored in the cloud [1]. These measures offer data as well as resource access control, and user and device authentication, including data confidentiality. Data is encrypted using cryptography, while it is protected from hackers using steganography. Key, data encryption and decryption for plaintext are accomplished in cryptography. Modern cryptography employs two kinds of keys, public and private, and requires complex mathematical calculations, making it the most secured approach. DNA cryptography [2, 3] is a modern, rapidly developing cryptographic method. The primary goal of this technique is to encrypt the plaintext and protect it in the digital DNA format. With the usage of OTP keys as well as its size, DNA cryptography [4, 5] offers better data secrecy than the current techniques. Additionally, there is less security because the methods currently in use used the same pattern and key generation strategies for all types of data. The idea of DNA computing [6, 7] is crucial in the area of computer security since it is now thought to be a more effective and unbreakable cryptographic method.
More than 100 lightweight encryption techniques have been presented as an alternative to the traditional techniques. The memory size, throughput or performance, and power consumption of IoT devices, and flexibility on various platforms for an IoT device present several challenges for the research and development of lightweight cryptographic algorithms. The data security level shouldn’t be lowered by any of this. The development and application of these algorithms nonetheless continue forward, but it is still difficult to discover algorithms that are appropriate for the particular requirements of IoT applications. In order to meet the computing requirements of IoT devices, it is also necessary to build a lightweight as well as flexible encryption approach [1, 8] that is based on using substitution as well as transposition as straightforward and effective logical operations to encode and decode data. Additionally, the lightweight encryption method must employ a straightforward procedure for creating keys [9, 10] that makes it challenging for attackers to crack them. Furthermore, using a lightweight, flexible encryption technique with variable data block sizes [11, 12] guarantees that system development will be more versatile to accommodate IoT devices with a variety of memory sizes. It will be much harder for adversaries to crack the updated DNA-based lightweight encryption technology because it creates keys randomly. In addition, the length and fundamental structure of DNA sequences allow for the logical and algebraic manipulation of data. These tasks must be completed in order to utilize the limited resources of IoT devices and avoid delays in real-time IoT applications. They also take a little amount of processing time. In this work, data hiding with respect to DNA sequence is referred to as modified DNA based cryptography.
The major contributions are:
• A modified DNA algorithm is proposed in this work for encryption with optimal key generation using deep learning model.
• A deep maxout model is used for generating the key for the encryption process, where the performance is ensured to be high in predicting the optimal key by tuning the model weights.
• Proposing a new hybrid optimization algorithm termed as FAAO algorithm for tuning the optimal weights of the deep maxout.
The organization of the paper is as follows: Section 2 discusses a literature review of existing DNA based cryptography system. Section 3 offers the proposed MDNAC. Section 4 provides the encryption using the modified DNA algorithm. Section 5 offers key generation using a deep maxout classifier. Section 6 provides the FAAO algorithm. Section 7 provides the validation part of MDNAC. Section 8 gives a conclusion.
2 Literature Review
In 2021, Mohammed Abbas Fadhil Al-Husainy et al. [13] developed a flexible, light-weight encryption method with robust, easy substitution, as well as transpose procedures to encrypt and decrypt data that is compatible with IoT devices’ limited computing power. By adopting a variable block size, the suggested method is made more adaptable to be used on numerous IoT devices with diverse memory capacities. Additionally, the DNA sequence is used to produce the randomized encryption keys that are challenging for hackers to crack.
In 2019, M. Indrasena Reddy et al. [14] suggested a DNA-based cryptographic system. The suggested method is based on modeling common DNA transcription, encoding, and translation mechanisms. Some inverse processes have also been employed for data encryption and decryption. In addition, likewise for the encryption/decryption techniques, it focuses on the CMDB. As a result, a BAMNN is used for storing information on the memory storage by identifying and recovering the pair of keys being used and using them to process randomized vital generation information.
In 2020, Pramod Pavithran et al. [15] presented a new cryptosystem depending on the idea of finite automata and DNA cryptography. The system consists of three components: sender, receiver, and key pair generator. Based on the characteristics of the receiver, the sender creates a secret key with a 256-bit DNA base that is used to encrypt data. Then, to increase the cypher text’s security, a Mealy machine that is generated at random is used to encode the DNA sequence.
In 2019, Md. Rafiul Biswas et al. [16] created a methodology for DNA cryptography assisted dynamic processes, i.e. “dynamic DNA encoding” as well as a dynamic sequence table. DNA base patterns are randomly applied to 256 ASCII characters in order to create the dynamic sequence database. In order to achieve dynamism, DNA base sequence locations are iteratively rearranged while adhering to a numerical series. An NCBI bank genome sequence and a numerical sequence are also utilized to build dynamic DNA encoding, which dynamically specifies the number of DNA bases required to assemble the encrypted text of every two chunks.
In 2019, SayantaniBasu et al. [17] developed a method depending on central dogma of molecular biology (CDMB) for encryption/decryption techniques, simulating genetic coding (converting from binary to DNA bases), transcription (converting DNA to mRNA), as well as transcription (converting mRNA to protein), and the opposite processes to enable encryption/decryption, respectively. It is assumed that all inputs come in the format of 16-bit blocks. The outputs from each block might be concatenated to produce the final encrypted text in the form of protein bases.
In 2021, NabarunNandy et al. [18] devised a method that minimizes the size of a text file and adds an additional layer of protection by using random key generation, assigning special codes to the most popular colors, and encrypting a variety of repeated colors. It is also less likely that the data will be recognized as an image when DNA elements A, T, G, and C are present. In this research, the text files are sent over an unsafe network
In 2022, Mona M. Elamir et al. [19] offered a unique idea for merging DNA strands obtained from encoded medical images and reporting to raise the security level across IoT networks, together with notions of DNA cryptography as well as a cryptosystem-assisted RSA approach. With this technique, high-quality images were successfully replicated. This proposed cryptosystem illustrated the viability of privacy and security in network security, particularly for e-healthcare employing an IoT system to promote medical collaboration and safely manage patient data across hospitals.
In 2019, Siyamol Chirakkarottu and Sheena Mathew [20] developed an innovative technique for encrypting medical images. The goal of the work was to propose an innovative, powerful, secure, and effective encryption method for medical images. It can also be used with any type of medical image, regardless of the storage format. The suggested approach swaps the pixels in the image using a pseudorandom number generator based on a 2D Zaslavski map. The concatenated image is encrypted using DNA encryption. Table 1 illustrates the features and challenges of existing DNA based cryptography systems.
Table 1 Features and challenges of an existing DNA-based cryptography system
| Author [Citation] | Methods | Features | Challenges |
| Mohammed Abbas Fadhil Al-Husainy et al. [13] | Lightweight cryptography system | Secured IoT data over the real-time attacks | Faced a number of difficulties as a result of the implementation’s size, speed, and power usage |
| M. Indrasena Reddy et al. [14] | WOA model | Allows for secure data storage and the encryption and decryption of massive volumes of data. | Gives a challenging slow convergence speed, low precision and falling into local optimal value easily. |
| Pramod Pavithran et al. [15] | Three entity based cryptosystem | Protect the system against various security attacks. | Spend a lot of time creating and retrieving secret keys. |
| Md. Rafiul Biswas et al. [16] | Dynamic mechanism based asymmetric cryptosystem | Increases data secrecy level | DNA calculations have not developed sufficiently. |
| Sayantani Basu et al. [17] | BAMNN | Capable of conserving memory by repeatedly memorizing and producing the pairs of keys | Reverse translation presented a substantial challenge since it involves a one-to-many mapping between a protein base as well as a 3-mRNA. |
| Nabarun Nandy et al. [18] | Random key generation and special code assignment algorithm | Encryption quality gets improved | The algorithm’s strength is highly competitive. |
| Mona M. Elamir et al. [19] | RSA model | Facilitate the secure handling of medical data across hospitals by the medical team | Data transfer is slow since there are many people involved. |
| Siyamol Chirakkarottu and Sheena Mathew [20] | 2-DZaslavski map | The suggested encryption method is resistant to differential attacks. | Execution time is high |
In 2023, AL-Shargabi [21] introduced a lightweight encryption approach that made use of the random structure of the DNA sequence to generate an encryption secret key. In order to overcome the limited processing and storage capacity of IoT devices, we developed the modified DNA-based lightweight encryption technique, which is based on the DNA sequence.
In 2023, Sharfuddin [22] described a novel method dubbed F-DNAES, which improves data security in cloud-based systems by fusing DNA cryptography with the advanced encryption standard (AES) algorithm. The suggested approach is centered on protecting data in transit and while it is being stored on cloud servers. F-DNAES offers a very safe and effective solution by combining the strength of AES encryption with the special qualities of DNA, such as its stability and storage capacity.
2.1 Review
Numerous challenges were encountered by the lightweight cryptography system due to the implementation’s size, speed, and power consumption [14]. The WOA model [14] gives a challenging and slow convergence speed, low precision and falls into the local optimal value easily. Three entity-based cryptosystems [15] spend a lot of time creating and retrieving secret keys. DNA calculations [16] have not developed sufficiently. Reverse translation [17] presented a substantial challenge since it involves a one-to-many mapping between a protein base as well as a 3-mRNA. Random key generation and special code assignment algorithm [18] strength is highly competitive. In the RSA model [19], due to the large number of participants, transmission of information is slow. Execution time is high in the 2D Zaslavski map [20]. DNA techniques beat traditional approaches in terms of time complexity, according to a simple comparison of conventional and DNA encryption systems that takes into consideration parameters like security level and time complexity. When compared to the existing DNA-based encryption methods, the modified DNA-based cryptography method proposed in this paper offers a higher level of security due to its added shifting operations during encryption and the effect of data block size.
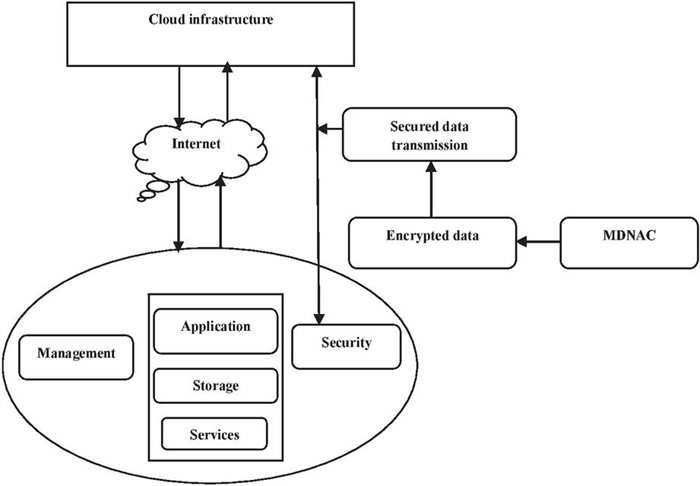
Figure 1 MDNAC based cloud security model.
3 Modified DNA-based Cryptography in the Cloud (MDNAC)
The transmission of hosted services, such as software, hardware, and storage, through the Internet, is known as cloud computing. Organizations of all sizes now use cloud computing almost universally, frequently as a component of a hybrid/multi-cloud IT architecture because of the advantages of rapid deployment, flexibility, cheap upfront costs, and scalability. The technology, rules, procedures, and services that safeguard cloud data, applications, as well as infrastructure against dangers are referred to as cloud security. The characteristics of DNA in the cloud are the organism’s genetic information necessary for its development and operation is contained in a molecule called DNA. Because of complementary base pairing, the sequences on one strand of the strand and another strand of the strand are chemically matched. Figure 1 shows the MDNAC-based cloud security. The proposed biologically inspired cryptosystem is the process of encryption and decoding using computer and biological processes. Due to its enormous storage and security capabilities, bio-inspired cryptography is a growing technique for data storage as well as security. DNA molecules could be paralleled, allowing for efficient energy consumption and increased data and storage densities. Information can be encrypted using modified DNA using numerous combinations of the four nucleotide bases and ML techniques to produce impossibly huge numbers. The core idea is based on a proper description of how biological encoding, transcription, and translation procedures were first identified and might be coupled for encryption during modified DNA encryption. Using a sample input; Figure 2 depicts the full encryption and decryption process. In this method, the input is equal to or below 16 bits and thus the process to inputs above 16 bits can be extended. Additionally, the cipher text which was subjected to the receiver, it is challenging to train the encoded format of value, and hashed protein values , and finalized key value formed using the deep maxout classifier with optimal tuning of weight .
Figure 2 Overall encryption and decryption procedure using sample input.
3.1 Cryptosystem
The existing system’s security and defense against attacks on important data and information depend on cryptography. Most likely, every modern cryptosystem consists of:
• Plain text: The first readable document to be converted into encrypted text is recognized to be the plaintext.
• Cipher text: Cipher text is the term used in cryptography to describe the process of converting an actual message into an unreadable message before transmission. This is a message that was obtained after some sort of plain text action that was encrypted.
• Key generation: A group of characters produced using key generation technique. The government utilizes asymmetrical key cryptosystems along with private keys, whereas symmetrical key cryptosystems use a single key for both encryption as well as decryption.
• Symmetric key cryptography: One of the existing and most well-known forms of encryption is symmetric encryption. The text signal modifies the text in a certain way by using one secret key, which might be a word, a number, or merely a collection of symbols. Moving a few letters around in the alphabet’s order might be sufficient to accomplish this. Every message utilizing this key could be encrypted or decrypted, even if neither the sender nor the recipients are aware of it.
• Asymmetric key cryptography: If secret keys are shared over a large network like the Internet, it might be problematic to ensure that they do not end up in the wrong hands. The data could be decrypted by someone who knows the secret key. One response is asymmetric encoding with two keys – a one key pair. A public key is willingly offered to anyone who might want to send a message. The personal key is then kept secret to ensure that only the recipient may access it next.
• Encryption: A way of converting plain text into cipher text is known as encryption. Cryptography uses the encryption technique to transmit private data via a socially anxious channel. Two components make up the encryption process: key as well as encryption method. The encryption technique infers the encryption. Intended receiver-side encryption is used for this.
• Decryption: Decryption is a technique for inverse encryption. The process involves changing plain text into encrypted text. The first signal is received using the receiver-side decryption procedure in cryptography. This cannot be in readable format. A method for decryption as well as a key is needed for the decryption process. The decryption method used is implied by the decryption process.
3.2 Encryption Using a Modified DNA Algorithm
In our research work, a modified DNA algorithm is employed for encryption process. The steps followed in the modified DNA algorithm is given below:
(1) Transform the input plain text into the corresponding binary values.
(2) Separate the plain text into four segments of equal size: .
(3) Create a random key with a length equal to , wherein n is the length of the plain text .
(4) Split the key into four equal parts: .
(5) Perform Kronocker product with key and then perform XOR operation. For example: with with with , and with ; here is the Kronocker product key.
(6) Concatenate the text after XOR operation.
(7) Convert the plain text into the DNA bases by performing transformation operation.
(8) Divide the plain text into ASCII values based on the DNA ASCII table, and apply decimal values.
(9) Binary values should be converted from decimal values.
(10) Replace binary values with the DNA bases.
(11) To create the final encryption text, acquire the values and replace those with the DNA ASCII table’s replaceable characters. After applying the ASCII, the key will be generated.
3.3 Key Generation Using the Deep Maxout Algorithm
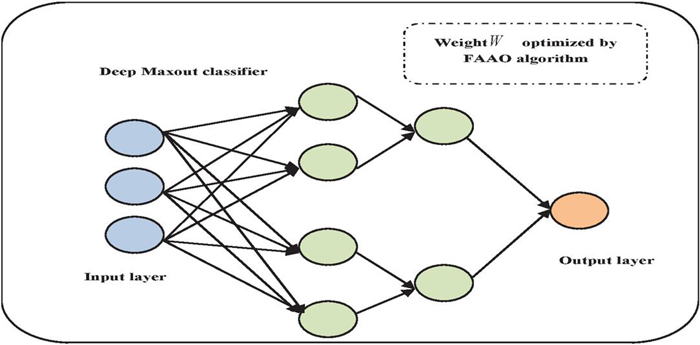
The key generation will be done using the deep maxout algorithm. For optimal key generation, the weight function of deep maxout will be fine-tuned by the proposed FAAO algorithm. Figure 3 illustrates the deep maxout classification process. Additionally, tests demonstrate that the deep maxout networks function effectively without dropout and are simple to improve without the requirement for pre-training.
Training phase: In the training phase based on the input data (as per the dataset), key corresponding to each data will be trained.
Testing phase: In the testing phase, depending on the training value the key will be generated.
Figure 3 Deep maxout classification for key generation.
3.3.1 Deep maxout classifier [23]
The input given to the DMN is . The maxout function is used by many layers of DMNs to accomplish hidden activations. All hidden units were divided at the discontinuity group layer of a maxout network. In this, is the total unit groups, and is the total units for each group. The maxout function is applied to each unit group to produce the activations for such a layer. Here, the output of deep maxout classifier is defined in Equation (1), here is depicted by linear pre-activation value. In this circumstance, is subjected to max pooling by the maxout function.
| (1) |
where, .
The maximum value within every group is considered to be lth layer in this case. Several maxout layers could be successively connected, and the softmax layer can then be added to create a DMN. The complete DMN might be pre-trained using layer-wise stacking AE (autoencoders) related to the maxout layers. The optimally tuned DMN weight is shown as and is determined by FAAO.
3.3.2 Feedback assisted Archimedes optimization (FAAO) algorithm for fine tuning deep maxout weight
AOA’s robustness stems from its ability to solve optimization issues by generating objective function values with minimal error, making it both simple to use and possessing few control parameters. In order to prevent being stuck in less-than-ideal settings, the AOA algorithm also keeps the flexibility to modify its pool of potential solutions. We developed a FAAO algorithm [24] for fine tuning the deep maxout weight . In this proposed algorithm, the position update of Archimedes in the exploration phase is done by the feedback artificial tree (FAT) algorithm [25]. The advantages of FAT are used to solve a large range of optimization problems. It does not require continuity and differentiability for the optimization. The following is a breakdown of the mathematical stages suggested by the FAAO.
Initialization: Using Equation (2), all object positions are initialized, where, indicates the ith object in an object population, indicates lower limit of search space, and indicates the upper limit of the search space. In Equation (3), for each ith object, the volume as well as density was initialized, where indicates the d-dimensional vector. Furthermore, in Equation (4), the ith object acceleration is initialized. In this step, the beginning position is evaluated, and the objects with highest fitness values are selected. Setting: , , , .
| (2) | |
| (3) | |
| (4) |
Update density and volume: Using Equation (5), the density and volume in object are updated for iteration , where indicates the volume/density with finest object, and indicates a uniformly distributed random integer.
| (5) |
Step 3: Proposed transfer operator and density factor: Following the initial collision and after some time has passed, the objects attempt to achieve equilibrium. This is achieved in AOA via the transfer operator , which changes searching from exploration to exploitation as shown in Equation (6), where increases slowly until it become 1, indicates the iteration number, and indicates the maximum iterations. Equation (8) was used to calculate the density reduction factor , and it decreased over time, similar to how it helps AOA with its local-to-global search, where decreased with time, and allows convergence in a best region which was already identified.
| (6) |
As per the FAAO, transfer operator is defined in Equation (7), where .
| (7) | ||
| (8) |
Exploration phase: If , collision among objects occurs, then is chosen as the random number, and the object’s acceleration is adjusted per iteration utilizing Equation (9), where , , indicate the density/volume/acceleration of object .
| (9) |
Exploitation phase: If , no collision occurs amongst the objects. In the exploitation phase, updating the acceleration of the object for iteration is gained by utilizing Equation (10), where indicates the best object acceleration.
| (10) |
Normalize acceleration: To determine the percentage of variation using Equation (11), the normalized acceleration is employed, where , indicates the normalization limit amongst [0.9, 0.1], and indicates the percentage of expected variations.
| (11) |
Proposed position update evaluation: (Exploration phase update). In Equation (12), here is constant number which equals 2, the ith position of the object for the following iteration is determined if .
| (12) |
As per the FAAO, the Archimedes position update is done via FAT which is expressed using Equation (3.3.2), where indicates the branch position.
| (13) |
In Equation (14), the position update of exploitation is done if , where indicates the constant number which equals 6, and indicates the rise in time, indicated as a directly proportional operator which is determined as . Flag is updated using Equation (15), where .
| (14) | |
| (15) |
Cycle crossover operation [26]: By filling each slot with a component from a separate parent, this operator aims to create an offspring from the parents. The following CX operation steps are listed:
• Starting with the first gene of the first parent, the cycle shifts to the first gene of the second parent.
• Find the gene that is in the first place in the second parent, and then move on to the corresponding gene in the first parent.
• Shift vertically from the current gene of the first parent to the gene of the second parent
• Make sure the first gene from the second parent is the same as the first gene from the second parent. Moving on to step 4 if the answer is “yes”; step 5 otherwise.
• After going on to the gene of the first parent that matches the gene of the second parent, proceed to step 3.
• Repeat the previous steps to create the second offspring.
• Transfer the genes from the cycle of the first parent to the corresponding places in the first offspring.
• Transfer the appropriate genes from the cycle of the second parent to the second offspring.
• Transfer the leftover genes from second parent into the equivalent places in the first offspring.
• Transfer the unused genes from the first parent to the second child’s genome at the appropriate places.
• The genes that are existent in every offspring combine to create the subsequent corresponding offspring.
Algorithm 1: Pseudocode of FAAO algorithm
Input: Output:
Procedure of FAAO: (population size , maximum iterations and )
Object population initialization is done using Equation (2), Equation (3), and Equation (4)
Using the optimal fitness value, initial population is evaluated
Set
While do
for each object do
Update volume and density via Equation (5)
Update as per FAAO via Equation (7) and via Equation (8)
If then
Update acceleration via Equation (9)
Update normalization using the Equation (11)
Exploration phase position update is done using Equation (3.3.2) as per FAAO
Else
Update acceleration via Equation (10)
Update normalization using the Equation (11)
Update flap via Equation (15)
Exploitation phase position update is done using Equation (14)
end if
end for
Assign
end while
Return optimal weight
end procedure
4 Results and Discussion
4.1 Simulation Procedure
The modified DNA based cryptography (MDNAC) model was implemented in Python/Cloudsim. Here, the MDNAC was measured against the optimization models like, feedback artificial tree (FAT), Archimedes optimization algorithm (AOA), blue monkey optimization (BMO), COOT, butterfly optimization algorithm (BOA), shark smell optimization (SSO), whale optimization algorithm (WOA) [13] and lightweight encryption system (LES) [14]. It was also contradicted with the encryption methods like, DNA, Blowfish, RSA, AES and ECC with respect to encryption time, decryption time, memory utilization and so on.
Figure 4 Analysis of MDNAC vs. standard methods: (a) decryption time, (b) encryption time, (c) memory utilization.
4.2 Analysis of Optimization Methods in Terms of Encryption Time, Decryption Time and Memory Utilization
The analysis of MDNAC was ascertained over the established methods, including, FAT, AOA, BMO, COOT, BOA and SSO. Here, the MDNAC secured cryptosystem was estimated with wide range of metrics like decryption time, encryption time and memory utilization. Also, it was computed for assorted key length and the findings are shown in Figure 4. As intimated by the figure, the MDNAC exceeded the other common techniques in terms of decryption time, encryption time and memory utilization. The decryption time of the MDNAC for the 4.0th key length is 0.23, which is indeed the minimal value as if related to FAT 0.42, AOA 0.38, BMO 0.46, COOT 0.51, BOA 0.43, SSO 0.42, WOA 0.39 and LES 0.44, respectively.
Considering Figure 4(b), in the initial (0.0) key length all the algorithms obtained extremely low encryption time but when the key length increased the encryption time also rose. Nevertheless, the MDNAC attained the lowest encryption time in almost all the key lengths. Moreover, when the key length was adjusted to 2.0, the MDNAC offered an encryption time of 0.07, even though the FAT, AOA, BMO, COOT, BOA, SSO, WOA and LES attained an encryption time of 0.16, 0.14, 0.08, 0.18, 0.19, 0.17, 0.15 and 0.09, respectively. Similarly, in Figure 4(c) the memory utilized by the MDNAC is 10.8 (key length 3.5), which is superior to FAT (30.2), AOA (20.4), BMO (30.6), COOT (20.9), BOA (30.3), SSO (30.1), WOA (20.7) and LES (20.6), respectively. Therefore, the MDNAC has accomplished negligible encryption and decryption time along with it consumed lower energy than the traditional optimization methods, demonstrating its suitability for secure cryptosystems.
Figure 5 Analysis of MDNAC vs. conventional encryption methods: (a) decryption time, (b) encryption time, (c) memory utilization.
4.3 Analysis of Encryption Methods in Terms of Encryption Time, Decryption Time and Memory Utilization
The MDNAC is evaluated with respect to the extant encryption methods such as DNA encryption, Blowfish, RSA, AES and ECC in terms of decryption time, memory utilization and encryption time, and is represented in Figure 5. Here, the memory use is much lower than for standard schemes and reduces both the encryption and decryption times. In particular, the MDNAC attained the decryption time of 0.07 in the key length 2.5, which is preferable to DNA encryption which is 0.12, Blowfish is 0.18, RSA is 0.15, AES is 0.14 and ECC is 0.13, respectively. Moreover, the MDNAC maintained the lowest encryption time for the fourth key length at 0.24, whilst DNA encryption, Blowfish, RSA, AES and ECC gained the encryption times of 0.35, 0.41, 0.38, 0.42 and 0.32, respectively. Furthermore, the key length is set to as 1.0, the memory utilized by the MDNAC is 0.04, exceeding DNA encryption 0.06, Blowfish 10.2, RSA 0.09, AES 10.4 and ECC 10.1, respectively. Further, the MDNAC has justified its superlative performance by reducing the encryption and decryption times for a secured cryptosystem.
Table 2 KPA, CPA and brute force attack analysis on MDNAC over optimization and encryption methods
| Algorithm | KPA (s) | CPA (s) | Brute force (s) |
| FAT | 0.042768 | 0.031295 | 13.0949 |
| AOA | 0.04983 | 0.020613 | 13.9304 |
| BMO | 0.028733 | 0.002952 | 14.4055 |
| BOA | 0.009287 | 0.046288 | 11.4041 |
| COOT | 0.052873 | 0.019643 | 12.2084 |
| SSO | 0.008867 | 0.042127 | 11.1514 |
| WOA | 0.072834 | 0.028966 | 13.1228 |
| LES | 0.056572 | 0.007921 | 14.748 |
| DNA encryption | 0.076297 | 0.092735 | 14.2989 |
| Blowfish | 0.063868 | 0.192873 | 13.8792 |
| RSA | 0.165863 | 0.029867 | 12.3797 |
| AES | 0.018692 | 0.007868 | 13.2786 |
| ECC | 0.009879 | 0.018973 | 11.8683 |
| MDNAC | 0.002877 | 0.003558 | 10.9729 |
4.4 Attack Analysis on MDNAC Over Optimization and Encryption Methods
An attack against a cloud computing platform, storage platform, or hosted application in a platform as a service or software as a service model is referred to as a cloud attack. The KPA, CPA and brute force attack estimation on MDNAC was assessed and determined with other approaches such as FAT, AOA, BMO, BOA, COOT, SSO, WOA, LES, DNA encryption, Blowfish, RSA, AES and ECC. The comparative findings are summarized in Table 2. In particular, the MDNAC provides high security to the cryptosystem when the KPA attack occurred and the attack rating in the MDNAC is 0.002877, which means that the MDNAC offers higher authentication to the data than the others, such as FAT 0.042768, AOA 0.04983, BMO 0.028733, BOA 0.09287, COOT 0.052873, SSO 0.008867, WOA 0.0772834, LES 0.056572, DNA encryption 0.076297, Blowfish 0.063868, RSA 0.165863, AES 0.018692 and ECC 0.09879, respectively. Similar to the other CPA and brute force attack analyses, the MDNAC affords improved security to the cryptosystem and ensures that it is protected from attackers.
4.5 Statistical Analysis with Respect to Correlation
To manifest the preeminence of the MDNAC, a statistical assessment was performed over the current methodologies regarding mean, standard deviation, minimum, median and maximum case scenario analysis. The pertinent findings are summarized in Table 3. The cryptosystem is better protected when the correlation rate declines. Moreover, the MDNAC attained the correlation value for the mean case scenario of 0.23716, though the FAT is 0.252766, AOA is 0.251486, BMO is 0.264649, COOT is 0.329773, BOA is 0.25435, SSO is 0.300476, WOA is 0.389052 and LES is 0.272143, respectively. Moreover, the MDNAC predominance is exhibited.
Table 3 Statistical analysis with respect to correlation
| FAT | AOA | BMO | COOT | BOA | SSO | WOA | LES | MDNAC | |
| Mean | 0.252766 | 0.251486 | 0.264649 | 0.329773 | 0.25435 | 0.300476 | 0.389052 | 0.272143 | 0.237166 |
| Median | 0.207683 | 0.237876 | 0.248797 | 0.348893 | 0.228757 | 0.286356 | 0.378902 | 0.297734 | 0.202979 |
| Standard deviation | 0.056341 | 0.076193 | 0.076222 | 0.104424 | 0.092282 | 0.075027 | 0.085648 | 0.076083 | 0.11788 |
| Minimum | 0.187648 | 0.156768 | 0.165983 | 0.193868 | 0.138769 | 0.193868 | 0.254677 | 0.165246 | 0.097686 |
| Maximum | 0.337592 | 0.357697 | 0.386562 | 0.486787 | 0.397468 | 0.386562 | 0.486787 | 0.368989 | 0.298768 |
4.6 Convergence Analysis on MDNAC
The convergence evaluation on MDNAC is compared to FAT, AOA, BMO, COOT, BOA, SSO, WOA and LES, and the findings are illustrated in Figure 6. The suggested method had a higher correlation rate during iteration 0 than COOT and WOA, and as the iteration rose, the MDNAC correlation value gradually declined. Additionally, the MDNAC retained the lowest correlation rate of 0.10 in the 25th iteration, whereas the BOA, BMO, COOT, BOA, SSO, WOA and LES each had correlation rates of 0.15, 0.18, 0.20, 0.20, 0.16, 0.21, and 0.19, respectively. Therefore, the MDNAC obtained significant outcomes when compared to existing approaches, highlighting its functionality to offer a secure cryptosystem.
Figure 6 Convergence analysis of MDNAC versus standard methodologies.
| Methods | Time (s) |
| FAT | 0.188523 |
| ArchOA | 0.126838 |
| BMO | 0.203887 |
| COOT | 0.173803 |
| BOA | 0.158798 |
| SSO | 0.143015 |
| WOA | 0.092878 |
| LES | 0.10388 |
| MDNAC | 0.078215 |
4.7 Computational Time
Table 4 shows the evaluation of computational time for the suggested MDNAC model over traditional techniques such as FAT, ArchOA, BMO, COOT, BOA, SSO, WOA, and LES. The computational time for the suggested model has obtains lowest rates (0.0782) over the existing models. Thus, the adopted EC+DHUEAO technique has minimal time for computation.
4.8 Discussion
In this work, data concealment with regard to DNA sequence is referred to as modified DNA-based cryptography, or DNAC. The established techniques, such as FAT, AOA, BMO, COOT, BOA, and SSO, were used to determine the analysis of MDNAC. Additionally, the memory used by the MDNAC is 0.04, the key length is set to 1.0, and the memory exceeds the values for DNA encryption (0.06), Blowfish (10.1), RSA (0.09), AES (10.1), and ECC (10.1). When producing the key for an encryption process, the deep maxout model is employed. By adjusting the model weights, high prediction accuracy is guaranteed. It optimizes deep maxout’s optimal weights by adjusting them using the FAAO approach, a unique hybrid optimization strategy.
5 Conclusion
A modified DNA based cryptography (MDNAC) was developed in this paper which is defined as data hiding with respect to DNA sequence. The MDNAC steps were: encryption, decryption, and optimal key generation. Cryptography uses the encryption technique to transmit private data via a socially anxious channel. Two components make up the encryption process: a key and an encryption algorithm. For the encryption algorithm, a modified DNA algorithm was employed. In the decryption phase, the reverse operation is performed to get the plain text from the cipher text. The key generation will be done using the classification process called deep maxout classification. In this, the weight of the deep maxout is optimally tuned by the proposed feedback assisted archimedes optimization (FAAO) algorithm. Finally, the performance of the MDNAC was evaluated and the output was verified successfully. Because DNA sequences are complicated and difficult to manipulate or decode, DNA cryptography offers a high degree of security. The authors concentrated on these challenges in an effort to encourage researchers to use the diligent work that has been done thus far in the field of DNA cryptography research, as well as to make use of the advantages and analysis of current works and attempt to go over any barriers that may exist. Encryption used in cloud cryptography protects data kept in the cloud. Cloud cryptography incorporates a number of safeguards that provide an additional layer of security to protect data from breaches, hacking, and virus infection. The several DNA cryptography methods were contrasted by the writers. These factors would also assist future researchers in designing and refining DNA storage methods for more dependable and efficient safe data storage.
References
[1] Pandey, Gyan Prakash, Implementation of DNA Cryptography in Cloud Computing and Using Huffman Algorithm, Socket Programming and New Approach to Secure Cloud Data (August 7, 2019). Available at SSRN: https://ssrn.com/abstract=3501494 or http://dx.doi.org/10.2139/ssrn.3501494.
[2] Majumdar, A., Biswas, A., Majumder, A. et al. A novel DNA-inspired encryption strategy for concealing cloud storage. Front. Comput. Sci. 15, 153807 (2021). https://doi.org/10.1007/s11704-019-9015-2.
[3] Kumar S. Ray, Mandrita Mondal, “Review on DNA Cryptography”, 2019.
[4] JarJar, A. Two Feistel rounds in image cryptography acting at the nucleotide level exploiting dna and rna property. SN Appl. Sci. 1, 1411 (2019). https://doi.org/10.1007/s42452-019-1305-7.
[5] Thangamani, N., Murugappan, M. A Lightweight Cryptography Technique with Random Pattern Generation. Wireless Pers Commun 104, 1409–1432 (2019). https://doi.org/10.1007/s11277-018-6092-8.
[6] Zhang, Y., Wang, F., Chao, J. et al. DNA origami cryptography for secure communication. Nat Commun 10, 5469 (2019). https://doi.org/10.1038/s41467-019-13517-3.
[7] Varsha Kolate1, R.B. Joshi, “An Information Security Using DNA Cryptography along with AES Algorithm”, Turkish Journal of Computer and Mathematics Education, 2021.
[8] Maria Imdad, Sofia Najwa Ramli, HairulnizamMahdin, “Increasing Randomization of Ciphertext in DNA Cryptography”, (IJACSA) International Journal of Advanced Computer Science and Applications, Vol. 12, No. 10, 2021.
[9] Prema T. Akkasaligar and Sumangala Biradar (2020) Selective medical image encryption using DNA cryptography, Information Security Journal: A Global Perspective, 29:2, 91–101, DOI: 10.1080/19393555.2020.1718248.
[10] Prasanna Balaji Narasingapuram, M. Ponnavaikko, “DNA Cryptography Based User Level Security for Cloud Computing and Applications”, International Journal of Recent Technology and Engineering (IJRTE) ISSN: 2277-3878, Volume-8 Issue-5, January 2020.
[11] BaraaTareq Hammad, Ali Maki Sagheer, Ismail Taha Ahmed, Norziana Jamil, “A comparative review on symmetric and asymmetric DNA-based cryptography”, Bulletin of Electrical Engineering and Informatics Vol. 9, No. 6, December 2020, pp. 2484–2491 ISSN: 2302-9285, DOI: 10.11591/eei.v9i6.2470.
[12] Anand M, Anusha G, Aravind Kumar MJ, Bharath Kumar R, Hema Varna M, “Cryptography Based on DNA Analysis”, Perspectives in Communication, Embedded-Systems and Signal-Processing (PiCES) – An International Journal ISSN: 2566-932X, Vol. 4, Issue 12, March 2021.
[13] Mohammed Abbas Fadhil Al-Husainy, Bassam Al-Shargabi, ShadiAljawarneh, “Lightweight cryptography system for IoT devices using DNA”, Computers and Electrical Engineering, Volume 95, October 2021.
[14] M.M. Indrasena Reddy, D.A.P. Siva Kumar, D.K. Subba Reddy, A secured cryptographic system based on DNA and a hybrid key generation approach, Bio Systems (2020), https://doi.org/10.1016/j.biosystems.2020.104207.
[15] Pramod Pavithran, Sheena Mathew, SuyelNamasudra, Pascal Lorenz, “A novel cryptosystem based on DNA cryptography and randomly generated mealy machine”, Computers & Security Volume 104, May 2021.
[16] Md. Rafiul Biswas, Kazi Md. RokibulAlama, Shinsuke Tamura, Yasuhiko Morimoto, “A technique for DNA cryptography based on dynamic mechanisms”, Journal of Information Security and Applications Volume 48, October 2019.
[17] SayantaniBasu, Marimuthu Karuppiah, MitaNasipuri, Anup Kumar Halder, Niranchana Radhakrishnan, “Bio-inspired cryptosystem with DNA cryptography and neural networks”, Journal of Systems Architecture, Volume 94, March 2019.
[18] Nandy, N., Banerjee, D. and Pradhan, C. Color image encryption using DNA based cryptography. Int. J. Inf. Tecnol. 13, 533–540 (2021). https://doi.org/10.1007/s41870-018-0100-9.
[19] Elamir, M.M., Mabrouk, M.S. and Marzouk, S.Y. Secure framework for IoT technology based on RSA and DNA cryptography. Egypt J Med Hum Genet 23, 116 (2022). https://doi.org/10.1186/s43042-022-00326-5.
[20] Chirakkarottu, S., Mathew, S. A novel encryption method for medical images using 2D Zaslavski map and DNA cryptography. SN Appl. Sci. 2, 1 (2020). https://doi.org/10.1007/s42452-019-1685-8.
[21] AL-Shargabi, B., and Dar Assi, A. (2023). A modified lightweight DNA-based cryptography method for internet of things devices. Expert Systems, e13270.
[22] Sharfuddin, N., Anwer, F., and Ali, S. (2023). A Novel Cryptographic Technique for Cloud Environment Based on Feedback DNA. Int. J. Exp. Res. Rev, 32, 323–339.
[23] Yajie Miao, Florian Metze, and Shourabh Rawat, “Deep Maxout Networks for Low-resource Speech Recognition”, 2013.
[24] Fatma A. Hashim, Kashif Hussain, Essam H. Houssein, Mai S. Mabrouk, Walid Al-Atabany, “Archimedes optimization algorithm: a new metaheuristic algorithm for solving optimization problems”, Applied Intelligence, https://doi.org/10.1007/s10489-020-01893-z.
[26] Varun Kumar S, and Dr. R. Panneerselvam, “A Study of Crossover Operators for Genetic Algorithms to Solve VRP and its Variants and New Sinusoidal Motion Crossover Operator”, International Journal of Computational Intelligence Research ISSN 0973-1873 Volume 13, Number 7 (2017), pp. 1717–1733 © Research India Publications http://www.ripublication.com.
Journal of Web Engineering, Vol. 22_8, 1075–1100.
doi: 10.13052/jwe1540-9589.2281
© 2024 River Publishers